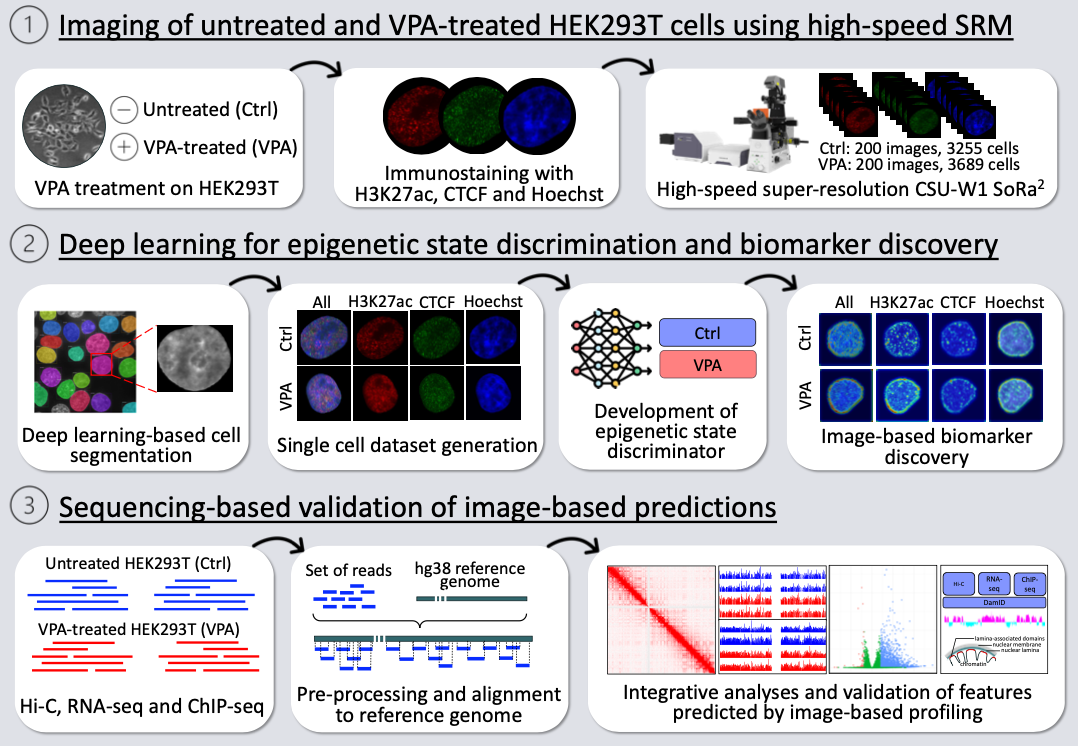
Abstract
Image-based profiling has been combined with machine learning for various tasks including cell classification and biomarker discovery. Epigenetic change within a cell nucleus can be captured by super-resolution microscopy (SRM), making it possible for image-based profiling. However, the low throughput of typical SRM has hindered its applications with machine learning. Here we employ a high-speed SRM called SoRa (Yokogawa CSU-W1) for image-based epigenetic profiling, and explore its applications to cell classification and biomarker discovery. As a model, HEK293T cells were treated with or without valproic acid (VPA), a histone deacetylase inhibitor to induce epigenetic change. The cells from each condition were stained for nuclear DNA (Hoechst), CTCF, and H3K27ac. 400 super-resolution images were obtained by SoRa within a day, and segmented into individual cell regions, yielding 6,944 cells in total. Using this large single-cell image dataset, we developed a deep learning method to discriminate the epigenetic states of cells (VPA-treated or not) with 99.6% accuracy. We also detected the image features that contributed to the discrimination as image-based biomarkers, suggesting the nuclear periphery as a hotspot of epigenetic change induced by VPA. To validate this image-based finding, we performed sequencing-based epigenomic analyses including Hi-C, ChIP-seq and RNA-seq. The integrative analysis with reported DamID data showed that genomic regions harboring epigenetic change were enriched in lamina-associated domains (LADs) at nuclear periphery, being consistent with the image-based finding. These results demonstrate the image-based epigenetic profiling combining SoRa and deep learning as a useful tool for cell classification and biomarker discovery.